|
|||||||||||||
|
|
|||||||||||||
|
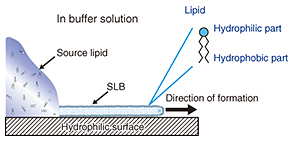
Feature Articles: Forefront Research on Bio-soft Materials Vol. 14, No. 8, pp. 18–22, Aug. 2016. https://doi.org/10.53829/ntr201608fa3 Pattern Formation of Supported Lipid Bilayer for Molecular ManipulationAbstractA supported lipid bilayer (SLB) is an artificial membrane formed on a solid support. We have developed an original technique for controlling the position of an SLB using a pattern fabricated on a solid surface. In this article, I report on two new results obtained using our technique; one involves the use of an SLB microarray for protein detection and the other a molecular gate device that can control a few individual molecules within an SLB. Keywords: lipid bilayer, self-spreading, molecule manipulation 1. IntroductionTen years have passed since we published our first paper describing the positional control of a supported lipid bilayer (SLB) and its application to microfluidic devices [1]. Our achievements in the early stages were detailed in a previous review [2]. I will focus on new developments in this article, especially on research using an SLB for transporting and manipulating biological molecules. Cell membranes are formed by various biological molecules. The main components of a membrane are lipids, which are molecules with both hydrophilic and hydrophobic parts. Therefore, a monolayer formed by lipid molecules has a hydrophilic surface and a hydrophobic surface. In aqueous conditions, the hydrophobic surfaces of two monolayers face each other to form a lipid bilayer, which is the basic structure of a cell membrane. Here, the lipid molecules can be obtained by extraction from natural products such as egg yolk and/or by chemical synthesis. A schematic drawing of self-spreading is shown in Fig. 1. First, the lipid molecules are placed on a hydrophilic solid surface. The solid surface is immersed gently in a buffer solution such as physiological saline. The lipid molecules form an SLB at the boundary between the lipid source and the solid surface, and it spreads across the solid surface over time. This phenomenon is driven by the self-organization process exhibited by the lipid molecules. Self-spreading occurs selectively on hydrophilic surfaces. Thus, when a solid has a hydrophilic/hydrophobic pattern surface, we can grow an SLB only on the hydrophilic surface. This is a very important technique for NTT Basic Research Laboratories in terms of accelerating our research in this field [1, 3].
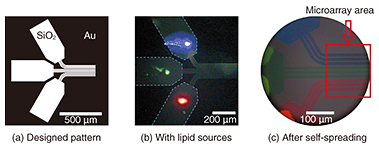
We observe SLBs using a fluorescence microscope. For the observations, we add a small quantity (~1%) of lipid molecules containing a fluorescent dye to the lipid source. Because the dye-conjugated lipid distributes homogeneously in the SLB, fluorescence images correspond to the shape of the SLB. 2. Fabrication of SLB microarrayA microarray is a two-dimensional array of biological molecules on a solid surface, which is useful for high-throughput medical screening. The best-known microarray is a deoxyribonucleic acid (DNA) chip. It can detect complementary DNA molecules in a sample solution on the basis of the hybridization of DNA fixed on a solid surface. Recent research has started to extend the technique to protein targets. This means proteins instead of DNA must be fixed on a solid surface. DNA is a stable molecule, and it maintains its activity when fixed on a solid surface. However, proteins are usually delicate molecules, and they often lose their activity as the result of fixation. One way to overcome this problem is to use an SLB as a cushion for proteins. If we are to develop the protein microarray on the basis of this idea, we must fabricate an SLB microarray with a high integration density. Our technique is useful for this purpose [4]. Our original proposed technique for fabricating a dense SLB microarray is shown in Fig. 2. Our strategy is to use pattern-guided self-spreading for the integration. A gold (Au) pattern fabricated on silicon dioxide (SiO2) is shown in Fig. 2(a). Different kinds of lipid sources, which show blue, green, and red fluorescence, are placed on the 500 × 250-μm areas (Fig. 2(b)) of the surface pattern on the solid. The sample is then immersed in buffer solution to initiate the self-spreading. As shown in Fig. 2(c), SLBs grow along the surface guide. At the end of the guide, we can obtain a linear pattern formed of 10-μm-wide SLBs with 5-μm intervals. The different colored dyes do not mix with each other because the lipid molecules are not soluble in aqueous media.
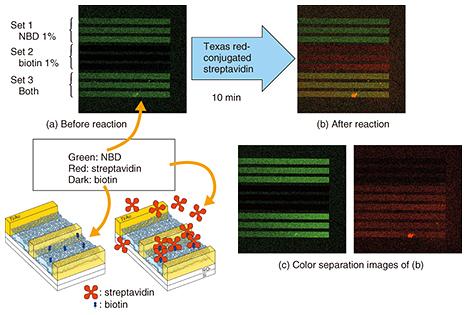
A technique for fabricating a dense SLB microarray was reported before our work. The technique is based on dropping vesicle solutions from different sources. One problem with the technique is that the droplets must not mix with each other during fabrication. Another problem is that of drying when the droplets are too small. The integration limit is thus determined by the droplet size. By contrast, SLBs are grown in a single buffer in our process. The integration limit is thus determined by the pattern size. This is the advantage of our technique, which yields an SLB microarray with a much higher integration density than the previous approach. 3. Protein detection by SLB microarrayWe applied our microarray to protein detection using the specific binding of streptavidin and biotin [4]. The results are shown in Fig. 3. The microarray has three different sets of SLBs, each formed of three lines. The first set contains 1% of nitro-benzoxadiazole (NBD)-conjugated lipid, the second set contains 1% of biotin-conjugated lipid, and the third set contains them both.
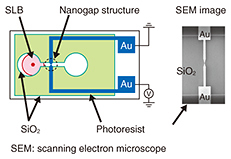
A fluorescence image of the microarray is shown in Fig. 3(a). In the initial stage, only NBD has green fluorescence. Thus, in Fig. 3(a) we observe green fluorescence in the first and third sets. Here we add a streptavidin solution. The streptavidin we used was labeled with red fluorescent dye, and thus, the red fluorescence must be observed from an area where the streptavidin is present. The fluorescence image of the microarray after the reaction is shown in Fig. 3(b). It can be separated into the green and red fluorescence images shown in Fig. 3(c). The red fluorescence, which corresponds to the position of streptavidin, is observed only in the second and third sets of SLBs, which include biotin-conjugated lipid. This confirms that our microarray works for protein detection. 4. SLB in nanostructuresWe can grow an SLB in the desired area by using a surface pattern as described above. Our next question concerns how narrow the area is in which an SLB can grow. To find the answer, we performed the following experiments [5, 6]. We designed the new structure shown in Fig. 4. It has a pair of gold electrodes with a 10–100 nm gap. The SLB is about 4 nm thick, and the gap is slightly larger than the thickness. The nanogap structure is much smaller than the resolution of an optical microscope. We therefore use a scanning electron microscope to observe the nanogap structure.
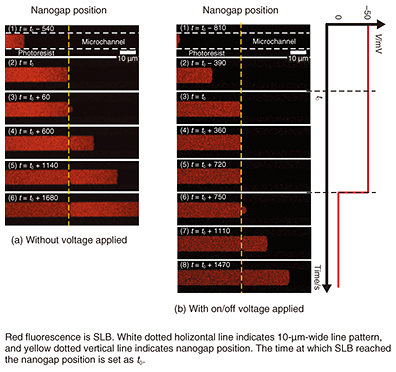
A self-spreading SLB passing through a nanogap is shown in Fig. 5(a). The SLB grows inside a 10-μm-wide line (1) and reaches the nanogap position (2). It continues to grow through the nanogap, which is located in the center of the line (3). The SLB keeps growing and reaches the other side of the nanogap (4–6). The most common way to fabricate SLBs is the vesicle fusion technique. However, it is difficult to determine if the SLB exists or not when we intend to fabricate the SLB in a nanostructure area such as the space in a nanogap structure. With our self-spreading method, it is clear that the SLB certainly grows on the nanogap area. 5. Manipulation of molecules inside SLBWe can use our nanogap structure as a pair of electrodes because they are made of Au metal. Next, we examined self-spreading when voltage was applied to the electrode. The self-spreading when –50 mV was applied between the electrodes is shown in Fig. 5(b) [5, 6]. The self-spreading behavior is no different from the example shown in Fig. 5(a) until the SLB reaches the nanogap (1–2). However, when the SLB reaches the nanogap (3), the SLB growth stops (4). After a certain period, the voltage is turned off (5), and the self-spreading restarts through the nanogap (6). The self-spreading after that is no different from that of the previous experiments shown in Fig. 5(a).
We explain the results with the following model. When an effective electric field is generated in the nanogap by the applied voltage, the field traps the molecule inside the SLB. This blocks the flow of the lipid molecules for SLB growth. In an electrolyte solution such as a physiological buffer, the electric field is only effective at nanometer-scale distances (called Debye length) from the electrode surface. In our experiments, we show that an effective electric field can be generated by using a nanometer-scale gap. If we reapply the voltage in Fig. 5(b)(8), we can again stop the growth. It stops the supply of the lipid molecules required for the growth by trapping them in the nanogap. Because the nanogap is very narrow and the effective electric field is applied to a limited area, we think that few molecules are trapped. This means we can control a macroscopic event such as self-spreading simply with a small number of molecules, which is analogous to a gate in a field-effect transistor [5]. The conditions for trapping molecules are determined by the gap distance and the Debye length, which varies depending on the electrolyte concentration in the solution. We checked the effect of these parameters by performing thorough experiments and revealed the validity of our model. 6. Future perspectivesI have described some of our recent results—especially those related to biosensing applications—that we have obtained as a result of our basic research activities. One of the goals is to develop a protein chip using our SLB microarray. A technique for manipulating molecules with nanogap electrodes can be further extended to single-molecule manipulation. It will constitute an original tool that will help NTT Basic Research Laboratories to undertake pioneering work in the new nano-bio research field. References
|
|||||||||||||