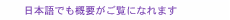|
|||||||||||
|
|
|||||||||||
|
Feature Articles: Forefront Research on Bio-soft Materials Vol. 14, No. 8, pp. 28–32, Aug. 2016. https://doi.org/10.53829/ntr201608fa5 Time-lapse Imaging of Neural Morphological Changes Relating to Cellular FunctionsAbstractThe human body is composed of trillions of cells. The association between the cells with specialized functions is important in the formation of the skeleton and organs, whose integration is vital for human activity. The nervous system plays a central role in effectively integrating the functions of the skeleton and organs. Here, we introduce research aimed at understanding the relationship between neural function and morphology in the early stages of apoptosis, which plays an important role in the process of neural network formation. Keywords: bioimaging, SICM, neuron 1. IntroductionThe human body is composed of several trillion cells. The association between the cells with specific functions plays a vital role in the formation of the skeleton and organs, whose integration is crucial for human activity. The nervous system plays a central role in integrating their functions so that they operate effectively. The nervous system is classified into two parts: the central nervous system, which is composed of the brain and spinal cord, and the peripheral nervous system, which is composed of the nerves throughout the body. In the nervous system, external stimuli detected by the peripheral nervous system are transmitted to the central nervous system. The central nervous system processes the information and sends the responses to the peripheral nervous system. For the nervous system to operate efficiently, a neurotransmission pathway network must be formed between neurons or between neurons and other cells. Therefore, it is important to understand the processes that form a neural network. There are two main processes involved in the formation of a neural network inside the brain. One is the neural maturation of individual neurons; this maturation provides functions for neurotransmission and the formation of a platform for neurotransmission, namely a synapse, between cells during neural network formation. The neural maturation processes can be classified into five stages based on established morphological criteria (Fig. 1). These consist of immature neurons lacking neurites (stage 1), neurons with multiple short neurites without established polarity (stage 2), and neurons that have achieved polarity and that possess neurites considerably longer than the rest, namely axons, which act as transmission paths (stage 3). The remaining neurites mature as dendrites with functions for receiving neural information from axons (stage 4). With further maturation, the axon and the dendrites elongate to form synapses among target cells (stage 5).
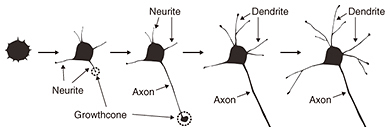
The other process is the formation of a neural network. One of the key biological events in this process is apoptosis. Apoptosis is defined as programmed cell death. In a developing nervous system, apoptosis occurs as a homeostatic mechanism to maintain cell populations in a neural network. It is a highly regulated and controlled process, and apoptotic cells are quickly eliminated without causing any damage to surrounding cells. In the neural network formation process, target cells secrete signals such as neural growth factors, which are essential for neural survival. Neurons that successfully receive the signals continue growing in the direction of the target cells. In contrast, neurons that fail to receive the signals are eliminated from the network formation process via several biological processes. If we are to understand this apoptotic process, we must investigate cellular functions at molecular and morphological levels. 2. Observation of biological samples by microscopyCellular morphologies are observed using some form of microscopy. Fluorescence microscopy is used to visualize biological samples by attaching fluorescence probes to them. This technique helps us to visualize the interaction between biological samples during biological events, although it is limited in terms of imaging morphological detail. Electron microscopy has been the conventional method for obtaining structural information about cells with higher resolution than optical microscopy, but it requires a fixed sample [1]. Therefore, it is impossible to obtain dynamic morphological images of living cells. One effective dynamic imaging technique, scanning probe microscopy [2, 3], and specifically, scanning ion conductance microscopy (SICM) [4], is expected to be a powerful tool for obtaining images of living cells (Fig. 2). This technique uses a glass pipette filled with an electrolyte that senses an ion current and feeds back its position relative to samples completely immersed in a liquid buffer containing electrolytes. Since the tip-sample distance is maintained at the radius of the pipette during the scan by keeping the ionic current constant, SICM enables stable and non-contact imaging of soft and sticky biological samples at a resolution of better than 100 nm. Because the application of external stimuli to living cells may induce cell death, SICM will allow us to image the morphological dynamics of living cells.
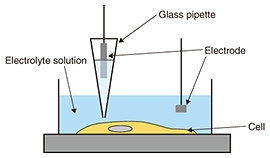
3. Neurons during apoptosisAs described above, apoptosis plays an important role in the neural network formation process. It exhibits a distinct set of the biological changes including apoptotic volume decrease, the formation of cell buds or membrane blebs, and phosphatidylserine (PS) translocation. The cell membrane is composed of the inner and outer leaflets of a lipid membrane. PS normally resides in the inner leaflet of the cell membrane and is translocated from the inner to the outer leaflet in the early stages of apoptosis. This PS translocation is considered to be a key biological event in relation to phagocytosis. As regards morphological changes during the early stages of apoptosis, the cell volume decreases, and membrane blebs are subsequently formed on the surface of the cell membrane. Despite the importance of these apoptotic morphological changes, no one has yet revealed the morphological dynamics at a resolution of better than a micrometer, or revealed the relationship between biochemical changes such as PS translocation and the cellular morphology. SICM is a suitable technique for investigating a series of these changes because it allows us to observe the morphology of living cells without any mechanical interactions between the probe and the sample surface with a high resolution. In this article, we discuss the morphological changes that occur in neurons in the early stages of apoptosis including apoptotic volume decrease and membrane blebbing. In addition, we explain our investigation of the relationship between membrane blebbing and PS translocation. Additionally, we use the results of these experiments to discuss the order of morphological dynamics during apoptosis and the relationship between biochemical and morphological changes [5]. 4. Bioimaging of neuronsWe investigated the morphological changes in apoptotic neurons by exposing cultivated neurons to staurosporine (STS), which is an apoptosis inducer. The morphological changes in a single neuron are shown in Fig. 3(a), and line scans for the SICM image indicated by the dashed line in Fig. 3(a) are shown in Fig. 3(b). In the first 120 min after exposure to STS, bulge-like structures known as membrane blebs were observed at the neuron (Fig. 3(a), white arrows). The number and size of the blebs increased with time. A graph of the volume change over time as a ratio of volume to initial volume, v/v0, shown in Fig. 3(c), was obtained from five neurons, and individual time-course images were obtained in an independent experiment. The v/v0 ranged from 0.6 to 0.8, meaning that the volume decreased from 20 to 40% in the first 120 min after exposure to STS. Once the volume reached its minimum value, it increased again, and membrane bleb formation was observed. The SICM results indicate that the time course of the morphological changes begins with an apoptotic volume decrease, followed by membrane blebbing.
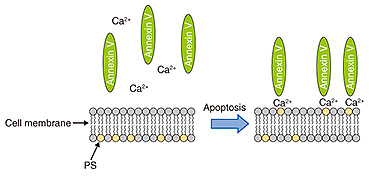
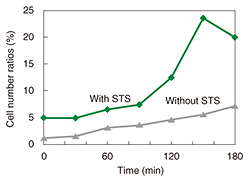
We used fluorescence microscopy to investigate the relationship between membrane blebbing and PS translocation by detecting the PS translocation with Annexin V, which binds to the PS residing in the outer leaflet of the cell membrane in the presence of calcium ions. Normally, PS exists in an inner leaflet of the cell membrane. Apoptosis induces the translocation of PS from the inner to the outer leaflet without changing the permeability of the cell membrane. This translocation can be detected via binding with fluorescently labeled Annexin V (Fig. 4). After the neurons were exposed to STS, we monitored the binding of the Annexin to the neurons (Fig. 5). The Annexin V-positive ratio without exposure to STS ranged from 0% to 10% during 180 min of observation, whereas the ratio with the exposure to STS increased more than 20%. This indicates that PS translocation induced by STS occurred around 120 min after exposure to STS.
These results obtained by SICM and fluorescence microscopy show that apoptosis induces a reduction in cellular volume and subsequent membrane blebbing. Moreover, the membrane blebbing had a similar onset time to the PS translocation. It has been difficult to observe the time series of morphological changes in living cells, especially that of soft and deformable structures. A non-contact imaging technique such as SICM provides us with the morphological details of neurons in the early stages of apoptosis. 5. Future perspectivesThe Molecular and Bio Science Research Group at NTT Basic Research Laboratories is focusing on fabricating a nanobiodevice based on biological systems. To achieve this, it is essential to understand how many biological systems are orchestrated inside organisms as well as to investigate individual biological materials and cells. The combination of conventional and new imaging techniques such as fluorescence microscopy and SICM will provide new insight into the relationship between individual biological events and applications to information technologies and medical technologies. References
|
|||||||||||